Sequencing DNA in our Extra Bedroom

MinION with flow-cell. The sensor is in the small window near the “SpotON” port. Video explanation of the principal of operation.
About a year ago, my girlfriend says, “I think we can sequence DNA in our extra room.” She showed me the website of Oxford Nanopore, which makes a USB DNA sequencer. We joked about it for a bit and that was that.
Until October, when I decided to buy her one for Christmas. I punched my credit card into their nice online store and waited for my shipping confirmation, which never arrived. Finally, I emailed them and found out I needed to join their “community.” At this point, I should mention that I’m not a biologist and I’m certainly not qualified to join a community of researchers. And they wanted to have a phone call. And this thing had better arrive by Dec. 25.
So I fessed up to my girlfriend (who has a PhD in genetics) and she wrote me a notecard with exactly what to say. I studied up. The day comes and I’m all nervous. The rep, I’ll call her Alice, phones and I proudly explain all the things I’m going to do, straight off my notecard.
Then Alice starts asking questions. I was not prepared for questions. Do I have a temperature controlled fridge? Duh, of course (in the kitchen)1 A freezer? Yes obviously2. A centrifuge and thermocycler? “Uh sure yeah I have that3.” Then Alice started asking questions about my research. Uh oh. I repeat something about bacterial colonies from my card but she isn’t buying it. I manage to get out that I understand this isn’t a spit-in-the-tube-and-done thing and that’s all I’ve got. She keeps pushing and I eventually admit that I’m really buying it for my girlfriend for Christmas. Apparently that’s okay, since once Alice finishes laughing she said that it will arrive by Dec. 25 but they don’t offer a gift-wrap service.
The box arrives, packed in some sweet dry ice stuff. Dec. 25 comes and I get a set of pipettes and a PCR machine.
The obvious thing to sequence is one of us but the MinION can only do 1-2 billion bases per run4; to have decent quality for a human you need more like 90 billion 5. We decide to swab our mouths before brushing our teeth and find out what’s in there. My girlfriend whips up some media, we plop our swabs in, and put the tubes in the $30 chicken egg incubator she found on Amazon. Two days later I’m informed that the cultures smell like “really really bad breath,” and that I “really ought to smell them for myself,” which I studiously avoid for the next two months.
With the help of a $80 genomic DNA extraction kit, we cut up the cells, filter all their bits out, and have genomic DNA ready to go. First we run a gel electrophoresis to prove to ourselves that we didn’t mess up the extraction too much.
At this step, all the real biologists out there are assuming I’m going to talk about quantifying the DNA to make sure we had the right concentration before blowing a $500 flow-cell (the consumable part of the MinION) on this. Yeah, that would be a lot of work and the line is pretty bright in the gel…
We open the fridge to get out the sequencer’s flow-cell and notice that everything is frozen. Oh %$@*#@#*. I set the fridge to “10” because that seemed like a good idea. Alice definitely isn’t going to buy my warranty-return story. Two days later we’ve got a new thermometer and are praying that 100 freeze-thaw cycles are, uh, totally fine.
Happily the MinION comes with a calibration program that doesn’t seem to notice our substandard storage: all green. At this point we discover that Oxford Nanopore helpfully sends everyone a set of sample DNA to run first. We decide that sounds like a really good idea.
The promotional videos for the MinION claim, “simple 10 minute setup.” About two hours later, we’ve done the library prep, and we’re pipetting into the device. There are lots of warnings on their webpage about how you really can’t let air into the thing (permanent destruction of the flow-cell, blah blah). So of course the first thing we do is introduce an air bubble. But it only covers half the sensor. I think it’s the most expensive 5µL of air I’ve ever seen.
It turns out the sequencer produces so much data the minimum requirements are a 1TB SSD and a quad-core CPU. My girlfriend’s laptop has 200 GB and a dual core, so that will have to suffice. We fire it up and it starts producing reads. We’re over the moon. 6-hours later the run finishes, but only 10% of the bases have been “called.” The way the system works is by reading tiny changes in electric current as the molecules pass through the nanopore. Apparently the signal processing is kind of hard because 2 days later it’s still going. I play Overwatch by myself.
Sequencing. You can see the 512 ports on the laptop screen. Approximately half are green (sequencing or waiting for DNA) and the other half are blue (air-bubbled).
The Oxford Nanopore website supports automated uploading and analysis of some datasets, including the calibration run. Our run produced 983 million (aligned) base pairs, which any professionals reading this are scoffing at, but I’m pretty sure Celera circa 2000 would have been impressed. I certainly am. The last time I did anything close to this, we tested PTC tasting with a gel electrophoresis that took all day and effectively sequenced 1 base. We prep and load our mouth-bacteria genomic DNA into the sequencer. We’re reusing the flow-cell to save money and it’s hard to load correctly. We’re all paranoid about air bubbles now, but there’s a ton of them in the little fluid pipes. Eventually we look at each other, shrug, and put the sample in. It goes nowhere.
The library prep includes adding “loading beads” to the sample which give the liquid a ghostly white color. You can see if your sample is on the sensor by tracking the movement of that color, and ours clearly was stuck on the top of the port. Eventually we searched the forums and found someone else incompetent enough to have the same problem, with a solution that can be summarized as, “pull some liquid out of a different port and hope.” It worked great.
The sequencer uploads data in realtime, so after about 20 minutes, we were looking at a report of what lives in our mouth. Good news: it’s all normal. Bad news: ew. Turns out that we’re hosting viruses that are preying on the bacteria we’re also hosting.
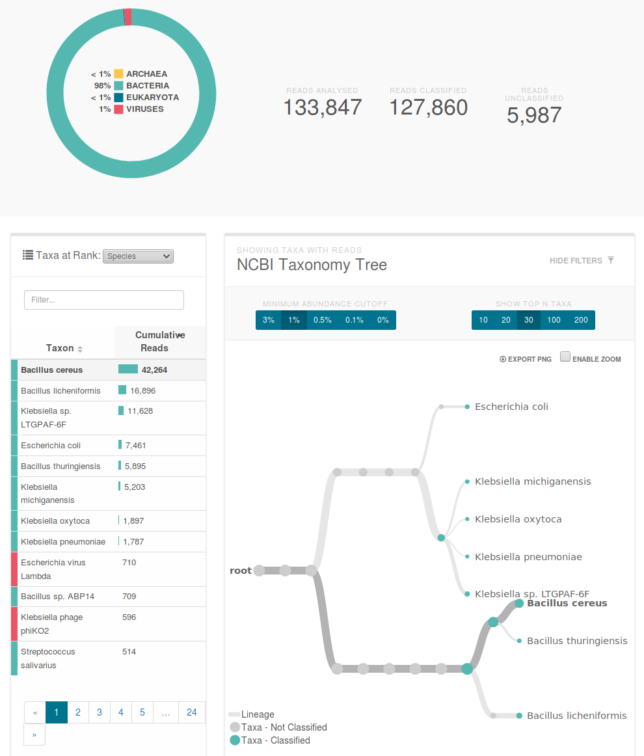
Classification of our data. The primary species are types of bacillus and klebsiella. You can see a klebsiella phage which is a virus that preys on the similarly named bacteria.
I can’t wait for the next time I get sick so I can confidently stride into the doctor’s office and inform them exactly what bacterial infection I have before throwing up on their table and finding out we massively contaminated the sample and I have a viral flu.
- it wasn’t and we soon acquired a new fridge
- yeah we bought a new one of those too
- hope that thing I just claimed to own isn’t too expensive
- in our hands; Oxford Nanopore claims 10-20 Gb now
- there’s a lot of variables here, go read this: Nanopore sequencing and assembly of a human genome with ultra-long reads
04/13/18